FROM TUMOUR BIOLOGY TO THERAPY DECISION IN THREE STEPS
The EndoPredict App illustrates the functional principle of the EndoPredict test, which is used for determining the prognosis of breast cancer patients. The test consists of the molecular finger print determined by the gene expression within the tumour tissue and is combined with the clinical status of the tumour. In context with other patient´s data, the test result assists the treating physicians and the tumour board in their decision as to whether chemotherapy is necessary or not.
The EndoPredict App answers the following questions:
1. How do the various genes in the tumour influence the prognosis?
2. How do the molecular fingerprint and the clinical data of tumour size and nodal status contribute to the EPclin score?
3. What is the benefit a patient can expect from the chemotherapy?
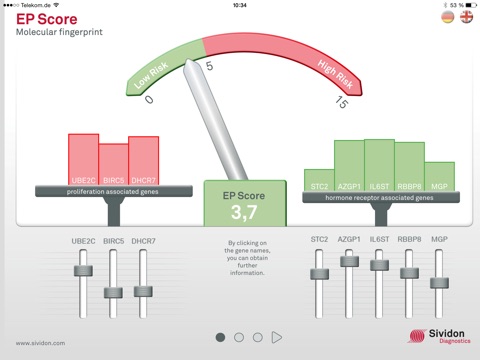
1. EP score
- Description and influence of eight disease-relevant genes
- Calculation of the EP score, the “molecular fingerprint”
►Possibility to simulate the influence of the individual genes by dragging the slider
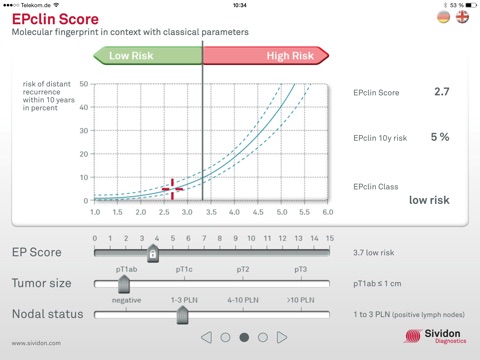
2. EPclin score
- Calculation of the EPclin score by adding clinical data about tumour size and nodal status to the EP score
- Statement on the risk of relapse within the next ten years
- Assignment to the high-risk or low-risk group of relapse (indicating chemotherapy/no chemotherapy)
►Dragging the slider gives an idea about the impact of the individual components on the overall test result.
►The EP score slider can be varied by double-clicking the lock symbol for the EP score.
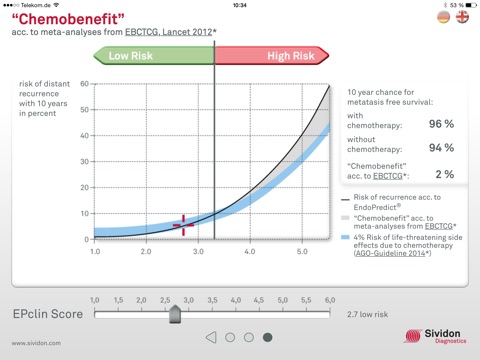
3. Chemobenefit
- Calculation of the “Chemobenefit” according to EBCTCG (Lancet 2012): Chemotherapy can reduce the risk of a metastasis by around one third
- Visualization of the potential benefit of a chemo therapy and its life-threatening adverse effects
►Dragging the slider allows the user to obtain the statistically determined Chemobenefit for respective EPclin scores.